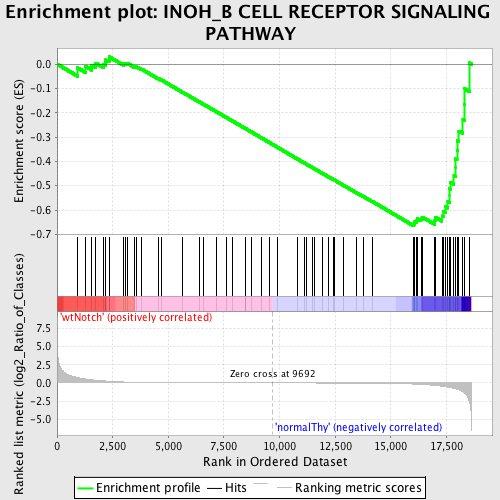
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | INOH_B CELL RECEPTOR SIGNALING PATHWAY |
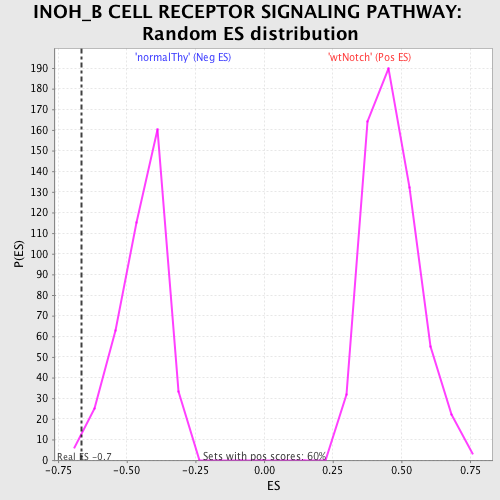
| Enrichment Score (ES) | -0.6654678 |
| Normalized Enrichment Score (NES) | -1.4952252 |
| Nominal p-value | 0.012437811 |
| FDR q-value | 0.30847558 |
| FWER p-Value | 0.999 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NFKBIB | 2260167 | 928 | 0.741 | -0.0158 | No | ||
| 2 | PIK3R2 | 1660193 | 1278 | 0.547 | -0.0094 | No | ||
| 3 | GRB2 | 6650398 | 1561 | 0.435 | -0.0046 | No | ||
| 4 | PRKCD | 770592 | 1727 | 0.382 | 0.0041 | No | ||
| 5 | HRAS | 1980551 | 2087 | 0.286 | -0.0020 | No | ||
| 6 | TRAF4 | 3060041 4920528 6980286 | 2176 | 0.267 | 0.0055 | No | ||
| 7 | ITPR3 | 4010632 | 2190 | 0.263 | 0.0170 | No | ||
| 8 | NFKB1 | 5420358 | 2339 | 0.229 | 0.0195 | No | ||
| 9 | MYD88 | 2100717 | 2345 | 0.227 | 0.0297 | No | ||
| 10 | SOS2 | 2060722 | 2998 | 0.130 | 0.0006 | No | ||
| 11 | FRS3 | 5890048 | 3059 | 0.124 | 0.0031 | No | ||
| 12 | PRKCZ | 3780279 | 3157 | 0.115 | 0.0032 | No | ||
| 13 | FADD | 2360593 4210692 6520019 | 3476 | 0.087 | -0.0100 | No | ||
| 14 | KRAS | 2060170 | 3552 | 0.081 | -0.0103 | No | ||
| 15 | PRKCA | 6400551 | 3810 | 0.064 | -0.0212 | No | ||
| 16 | MAP2K2 | 4590601 | 4558 | 0.036 | -0.0598 | No | ||
| 17 | SOS1 | 7050338 | 4675 | 0.034 | -0.0645 | No | ||
| 18 | GRB14 | 510035 3060369 | 5641 | 0.020 | -0.1156 | No | ||
| 19 | GRAP2 | 7100441 1410647 | 6397 | 0.013 | -0.1557 | No | ||
| 20 | IRS4 | 430341 1190450 | 6568 | 0.012 | -0.1643 | No | ||
| 21 | PIK3R1 | 4730671 | 7181 | 0.009 | -0.1969 | No | ||
| 22 | DOK1 | 1050279 2680112 5220273 | 7620 | 0.007 | -0.2202 | No | ||
| 23 | SHC3 | 2690398 | 7887 | 0.006 | -0.2342 | No | ||
| 24 | MAP2K1 | 840739 | 8448 | 0.004 | -0.2642 | No | ||
| 25 | NRAS | 6900577 | 8451 | 0.004 | -0.2642 | No | ||
| 26 | BCAR1 | 1340215 | 8752 | 0.003 | -0.2802 | No | ||
| 27 | PRKCQ | 2260170 3870193 | 9203 | 0.001 | -0.3044 | No | ||
| 28 | GAB2 | 1410280 2340520 4280040 | 9538 | 0.000 | -0.3224 | No | ||
| 29 | PIK3CA | 6220129 | 9883 | -0.001 | -0.3409 | No | ||
| 30 | CD19 | 6940692 | 10822 | -0.003 | -0.3913 | No | ||
| 31 | SRC | 580132 | 11138 | -0.004 | -0.4081 | No | ||
| 32 | PRKCE | 5700053 | 11223 | -0.005 | -0.4124 | No | ||
| 33 | SH2B3 | 4810451 4850128 | 11495 | -0.006 | -0.4267 | No | ||
| 34 | PRKCI | 5420139 | 11553 | -0.006 | -0.4295 | No | ||
| 35 | CRADD | 6770520 | 11908 | -0.007 | -0.4483 | No | ||
| 36 | IRS1 | 1190204 | 12178 | -0.009 | -0.4624 | No | ||
| 37 | GRB10 | 6980082 | 12405 | -0.010 | -0.4741 | No | ||
| 38 | TRAF5 | 3290064 | 12470 | -0.010 | -0.4771 | No | ||
| 39 | BLNK | 70288 6590092 | 12874 | -0.013 | -0.4982 | No | ||
| 40 | CD79A | 3450563 | 13468 | -0.018 | -0.5294 | No | ||
| 41 | SYK | 6940133 | 13759 | -0.022 | -0.5440 | No | ||
| 42 | SHC1 | 2900731 3170504 6520537 | 14167 | -0.028 | -0.5647 | No | ||
| 43 | LCP2 | 2680066 6650707 | 16038 | -0.152 | -0.6585 | Yes | ||
| 44 | MAPK3 | 580161 4780035 | 16052 | -0.154 | -0.6521 | Yes | ||
| 45 | ARAF | 6450270 | 16082 | -0.159 | -0.6463 | Yes | ||
| 46 | GRB7 | 2100471 | 16166 | -0.173 | -0.6428 | Yes | ||
| 47 | FYN | 2100468 4760520 4850687 | 16182 | -0.174 | -0.6355 | Yes | ||
| 48 | CRK | 1230162 4780128 | 16356 | -0.203 | -0.6355 | Yes | ||
| 49 | LAT | 3170025 | 16412 | -0.213 | -0.6287 | Yes | ||
| 50 | RELA | 3830075 | 16981 | -0.335 | -0.6438 | Yes | ||
| 51 | RAF1 | 1770600 | 16999 | -0.340 | -0.6291 | Yes | ||
| 52 | TRAF6 | 4810292 6200132 | 17300 | -0.454 | -0.6243 | Yes | ||
| 53 | LCK | 3360142 | 17374 | -0.493 | -0.6055 | Yes | ||
| 54 | PRKCH | 5720079 | 17442 | -0.520 | -0.5851 | Yes | ||
| 55 | TRAF2 | 5900148 | 17566 | -0.574 | -0.5653 | Yes | ||
| 56 | PIK3CD | 3060546 | 17647 | -0.614 | -0.5413 | Yes | ||
| 57 | VAV1 | 6020487 | 17649 | -0.615 | -0.5130 | Yes | ||
| 58 | GAB1 | 2970156 | 17684 | -0.634 | -0.4856 | Yes | ||
| 59 | CBL | 6380068 | 17838 | -0.744 | -0.4595 | Yes | ||
| 60 | SH2D2A | 2970594 | 17911 | -0.795 | -0.4267 | Yes | ||
| 61 | NFKBIA | 1570152 | 17914 | -0.797 | -0.3900 | Yes | ||
| 62 | CRKL | 4050427 | 17976 | -0.847 | -0.3542 | Yes | ||
| 63 | DOK2 | 5340273 | 17990 | -0.859 | -0.3153 | Yes | ||
| 64 | TRAF1 | 3440735 | 18060 | -0.950 | -0.2752 | Yes | ||
| 65 | TRAF3 | 5690647 | 18242 | -1.233 | -0.2281 | Yes | ||
| 66 | ITPR1 | 3450519 | 18312 | -1.399 | -0.1673 | Yes | ||
| 67 | NFKBIE | 580390 2190086 | 18321 | -1.432 | -0.1016 | Yes | ||
| 68 | CD79B | 1450066 3390358 | 18530 | -2.547 | 0.0046 | Yes |